Computational Analyses of Docosahexaenoic
Acid (DHA, C22:6, n-3) with Alzheimer’s
Disease-Causing Amyloid Peptide Aβ 1-42
Reassures Its Therapeutic Utility
Michio Hashimoto 1* , Shahdat Hossain 1,2 , Kentaro Matsuzaki 1 , Abdullah Al Mamun 1 ,
Hiroyuki Arai 3 , Osamu Shido 1
1 Department of Environmental Physiology, Faculty of Medicine, Shimane University, Izumoshi, Japan
2 Laboratory of Alternative Medicine and Behavioral Neurosciences, Department of Biochemistry and Molecular
Biology, Jahangirnagar University, Dhaka, Bangladesh
3 Department of Geriatrics and Gerontology, Division of Brain Sciences, Institute of Development, Aging and
Cancer, Tohoku University, Sendai, Japan
Received 4 April 2016; accepted 26 June 2016; published 29 June 2016
Copyright © 2016 by authors and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/Abstract
The accumulation of amyloid β peptide 1-42 (Aβ 1-42 ) masses in the brains of Alzheimer’s Disease (AD)
patients is associated with neuronal loss and memory deficits. We have previously reported that
oral administration of docosahexaenoic acid (DHA, C22:6, n-3) significantly decreases Aβ burden
in the brains of AD model rats and that direct in vitro incubation of DHA with Aβ 1-42 curbs the pro-
gression of amyloid fibrillation. In the present in silico study, we investigated whether DHA com-
putationally binds with amyloid peptides. The NMR solution structures of Aβ 1-42 were downloaded
from the Protein Data Bank (PDB IDs: 1Z0Q and 2BEG). The binding of DHA to Aβ peptides was as-
sessed by molecular docking using both a flexible and rigid docking system. Thioflavin T (ThT)
was used as positive control. The chemical structures of ThT and DHA were modeled and con-
verted to the PDB format using PRODRUG. Drug-like properties of DHA were evaluated by ADME
(Absorption, Distribution, Metabolism, and Excretion). DHA was found to successfully dock with
Aβ 1-42 . Computational analyses of the binding of DHA to Aβ 1-42 , as evaluated by docking studies,
further corroborated the inhibitory effect of DHA on in vitro Aβ 1-42 fibrillogenesis and might ex-
plain the in vivo reduction of amyloid burden observed in the brains of DHA-administered AD
model rats demonstrated in our previous study. These computational data suggest the potential
utility of DHA as a preventive medication in Aβ-induced neurodegenerative diseases, including AD.
* Corresponding author.
M. Hashimoto et al.
74
Keywords
Docosahexaenoic Acid, Alzheimer’s Disease, Amyloid Beta Peptide, Molecular Docking, In Silico,
Drug Design, Protein Data Bank
1. Introduction
Alzheimer’s Disease (AD), a progressive neurodegenerative disease, is the most common cause of dementia in
elderly people [1] [2]. Clinically, it is characterized by loss of memory, inability to learn new things, loss of
language function, a disturbed perception of space, inability to do calculations, and a host of other manifesta-
tions [3]. In AD, there is an overall shrinkage of brain tissue, whereas, it is microscopically characterized by
extracellular (neuritic plaques) and/or intracellular (neurofibrillary tangles) deposition of insoluble amyloid beta
peptides (Aβs) [4]. Aβ is a peptide with 42 or 43 amino acids that forms a part of the transmembrane domain of
the larger Amyloid Precursor Protein (APP), a transmembrane protein expressed in neurons and other brain cells
and is derived from the cleavage of APP by the β- and γ-secretase enzymes [5]-[7]. Defective clearance of Aβ
from aberrant cleavage of APP and other mechanisms results in its accumulation [8] [9]. Aβ monomers initially
polymerize into soluble oligomers and subsequently into larger insoluble fragments which precipitate as amyloid
fibrils [10]. The beta amyloid hypothesis suggests that AD is caused by the deposition of Aβ in plaques in brain
tissue and is the basis of a novel prevention and treatment method for AD [11]. Indeed, AD animal models can
be produced by brain ventricular infusion of Aβ 1-42 [12] [13]. Moreover, other fragments of Aβs, such as Aβ 1-40
[14]-[16] and Aβ 25-35 [17], have also been shown to assemble into amyloid fibers in vitro [16] [18] and have
been infused into rat brain ventricles to create AD animal models. Because AD has been shown to be associated
with decreased Docosahexaenoic Acid (DHA) levels [19] [20], numerous human trials are currently ongoing to
determine whether DHA is effective in the treatment and/or delaying the symptoms of AD [21] [22]. We also
reported that DHA decreases amyloid burden in the brains of Aβ-infused model rats [14] [15] and inhibits in vi-
tro amyloid fibrillation [16] [18]. These investigations highlight the potential for DHA as an excellent therapeu-
tic agent against Aβ-induced neurodegenerative diseases, including AD.
Previously, we showed that DHA inhibits Aβ fibril formation [10] [14]-[16] [18]. In this study, we investi-
gated the computational binding of DHA to Aβ peptides to further support the anti-Aβ fibrillation effect of DHA.
Because X-ray crystallographic structures of Aβs are not available, NMR structures were utilized in the present
investigation. We performed molecular docking studies, which are useful for identifying agents capable of inhi-
biting proteins responsible for disease pathology and exploring their possible binding modes. We used thiofla-
vin-T (ThT) as a positive control because it is currently used as a “standard” for selectively binding, staining,
and identifying amyloid fibrils both in vivo and in vitro [23]. We docked DHA and ThT (as ligands) onto Aβ 1-42
(as receptors) and compared their binding modes. We aimed to confirm the results of our previous studies,
showing the ameliorative effects of DHA and its potential as an AD therapy, in this computational analysis [10]
[13]-[16] [18].
2. Methods and Materials
2.1. Preparation of Thioflavin T and Docosahexaenoic Acid Models
DHA (CID: 445580) and ThT (CID: 16953) were downloaded from the PubChem database in sdf format. The
sdf files were submitted to the PRODRG server [24] to give energy-minimized structures of the ligands. Energy
minimization is performed to help the docking program identify the bioactive conformer from the local minima.
The two-dimensional structures of ligands are illustrated as ball and stick in Figure 1.
2.2. Analyses of Drug-Like Properties of the Ligands
Drug design essentially focuses on optimizing the binding interactions of ligands with their targets. However, a
compound with the best binding interactions does not necessarily make the best drug, as other factors are in-
volved. For example, a clinically useful drug must travel through the body to reach its target. Perfecting a com-
pound with good drug-target interactions is irrelevant if it has no chance of reaching its target. The factors that
M. Hashimoto et al.
75
Figure 1. The ligand molecules thioflavin T (ThT) and docosahexaenoic acid (DHA, C22:6, n-3).
determine whether a drug will reach its target in the body are termed the pharmacokinetic or ADME (Absorp-
tion, Distribution, Metabolism, and Excretion) properties. We used the free ADME/tox filtering tool (FAF-Drugs
3), which can predict whether the physicochemical properties of compounds are acceptable by applying several
filtering rules, including the well-known Lipinski’s rule of five (i.e., molecular weight > 500, logP or octanol/
water partition coefficient < 5, number of H-bond donors < 5, and number of H-bond acceptors < 10) [Lipinski
et al., 2001] [25]. The main properties computed by FAF-Drugs 3 are the number of rigid and flexible bonds,
topological polar surface area (TPSA) value according to Ertl et al. (2000) [26], number and maximum size of
system rings, and presence of unwanted chemicals or chemical substructures (using SMARTS searches) [27].
2.3. Analysis of the Amyloidogenic Regions in the Aβ 1-42 and Cross-Beta Aggregation
Sequence-based computational methods, namely FoldAmyloid [28], AGGRESCAN [29], and ProA [30], were
used to predict the amyloidogenic regions in Aβ 1-42 . Furthermore, analysis of the beta-aggregation propensity of
Aβ 1-42 was achieved by subjecting the primary amino acid sequence of Aβ 1-42 to the TANGO algorithm [31]-
[33], which is designed to predict cross-beta aggregation in peptides.
2.4. Preparation of Aβ 1-42 Receptor Models
The three-dimensional solution structures (3D NMR) of Aβ 1-42 (PDB IDs: 2BEG and 1Z0Q) were downloaded
from the RCSB Protein Data Bank (PDB) (
http://www.rcsb.org/) as receptors for ThT and DHA docking. 2BEG
is a homopentamer, namely composed of the A, B, C, D, and E monomers of Aβ 1-42 . Each monomer (A, B, C, D,
and E) of 2BEG comprises 10 coordinate models [9]. The coordinates of model 1 of the A monomer (A1), mod-
el 1 of the B monomer (B1), and model 1 of the C monomer (C1) were loaded in the Molegro virtual docker
(MVD) and split from the composite 2BEG PDB file. Accordingly, 1Z0Q is a homo 30-mer composed of 30-
monomers (A), each comprising an individual coordinate model. The A1 monomer was split from the composite
1Z0Q PDB file. At least two amyloid molecules are required to achieve the repeating structure of a protofila-
ment fibril. Therefore, the dimer (Figure 2) or the trimer (Figure 6) models were generated by RossettaDock
[34], as described previously [35]. Subsequently, the generated dimers (A1B1) or trimers (A1B1C1) were uti-
lized for ThT and DHA docking (both flexible and rigid docking).
2.5. Computational Analysis of Binding Sites in Aβ 1-42 Dimers or Trimers
The binding sites or pockets of the dimer (A1B1) and trimer (A1B1C1) were determined by GHECOM, which
detects grid-based pockets/binding sites on the surface/interior of the protein [36]. The program produces a
graph of residue-based “pocketness,” and the presence of binding sites was visualized using the 3D molecular
viewer Jmol [37].
2.6. Inter-Surface Interaction Site Analysis of the Aβ 1-42 Dimer (A1B1)
Protein-protein (the monomer-monomer A1B1 dimer) inter-surface interaction sites were analyzed by feeding
the A1B1 dimer to the cons-PPISP server [38], which predicts the residues that are likely to form the binding
site for neighboring proteins. The energy distribution along the protein-protein interface is not homogenous;
M. Hashimoto et al.
76
Figure 2. The monomeric structure files for Aβ 1-42 were prepared from their parent composite PDB files (2BEG and 1Z0Q).
Subsequently, dimers of Aβ 1-42 were formed in order to dock ligands (ThT and DHA). (a), (b) Monomer; (c), (d) Dimer.
certain residues contribute more to the binding free energy, called “hot spots.” Hot-point servers predict hot
spots in protein interfaces using an empirical model that incorporates a few simple rules consisting of occlusion
from solvent and total knowledge-based pair potentials of residues [39]. To further validate the presence of in-
ter-surface hotpoints on the dimer interface binding contacts, the dimer was fed to hot-point prediction servers,
including Knowledge-based FADE and Contacts (KFC2) [40].
2.7. Molecular Docking Simulations of ThT and DHA onto the Aβ 1-42 Dimer
The molecular docking simulations were performed using MVD [41] and PatchDock [42].
2.7.1. Molegro Virtual Docker (MVD)
MVD version 4.3.0 (free academic version), along with its graphical user interface (MVD tools), was utilized to
generate a grid, calculate the dock score, and evaluate conformers. The nonpolar hydrogen atoms were removed
from the receptor file and their partial charges were added to the corresponding carbon atoms. Docking was
performed by following the steps in the MVD user manual [41]. MolDock score of ligands were calculated dur-
ing docking. MVD performs flexible ligand docking with optimization of the ligand geometry during docking.
Therefore, bond angles, bond lengths, and torsional angles of the ligand are modified during the stages of recep-
tor-ligand complex generation.
2.7.2. PatchDock
PatchDock is a shape complementarity/geometry-based molecular docking algorithm. It is aimed at finding
docking transformations that yield good molecular shape complementarity. Such transformations induce both
wide interface areas and small amounts of steric clashes. A wide interface is ensured to include several matched
local features of the docked molecules that have complementary characteristics. The output of PatchDock is a
list of candidate complexes between the receptor and ligand molecules. The list is sorted according to geometric
shape complementarity score, approximate interface area of the receptor-ligand complex, atomic contact energy
(ACE) between ligand and receptor, and 3D transformation. Finally, the server provides an option to download
the ligand-receptor complexes in the PDB format. The PatchDocked ligand-receptor complex (of Rank 1) was
visualized using PyMOL [43].
3. Results
The predicted physical (ADME) properties of DHA and ThT, as evaluated by the FAF-Drug 3, are shown in
M. Hashimoto et al.
77
Table 1. The major difference between the properties of DHA and ThT was in the LogP (octanol-water partition
coefficient) value, which was >5 for DHA, but <5 for ThT. Thus, DHA violated Lipinski’s rule of five and this
may relate to the more hydrophobic acyl chains in DHA compared with those in ThT. Partition coefficients in-
dicate the distribution of drugs within the body. Hydrophobic drugs with high octanol/water partition coeffi-
cients are mainly distributed to hydrophobic areas such as lipid bilayers of cells. The number of rotatable flexi-
ble bonds helps a molecule to adopt 3D structure with a greater flexibility, while that of the rigid bonds performs
the opposite trend. Molecular polar surface area (PSA) is a very useful parameter for prediction of drug transport
properties. Polar surface area is defined as a sum of surfaces of polar atoms (usually oxygens, nitrogens and at-
tached hydrogens) in a molecule. This parameter is used to correlate very well with the human intestinal absorp-
tion, monolayers permeability, and blood-brain barrier penetration. Usually, the molecules with a polar surface
area of greater than 120 - 140 Å 2 tend to be poor in permeating cell membranes. The oral bioavailability scores
based on solubility in water (Sw) were 2.29 × 10 −5 and 2.14 × 10 −5 for DHA and ThT, respectively. The log of
these two values, i.e. Sw, was −4.64 and −4.67, respectively. This indicates that the water solubility of these two
compounds were comparable. Because of the Lipinski’s rules (LR) are based on physicochemical criteria for
physiological effect of drugs (ADME mainly) but does not rule out a possible direct interaction between a mo-
lecule and a protein outside the cellular context. Moreover, LRs are violated by numerous drugs or physiologi-
cally relevant molecules. Notably, the LR is a group of physicochemical properties used to evaluate the proba-
bility of a substance to become an effective drug. However, the conclusions that can be drawn from a docking
exercise are limited. If a docking is good and a ligand gets a good score, this just suggests that the compound
might have activity against a given target. Docking says nothing about physicochemical properties.
Aggregation-propensity analyses of the Aβ 1-42 peptides are shown in Table 2. The hot-spot analyses by
FoldAmyloid [28], AGGRESCAN [29], and ProA [30] revealed that the amino acid residues from Val24 to
Lys28/ Gly29 and those from Val39 to Ala42 were the most susceptible to amyloidogenesis.
Beta-aggregation propensity analysis also revealed that the amino acid residues from Lys16 to Asp23, and
those from Lys28 to Ala42, displayed the highest beta-aggregation tendency (Figure 3). These two regions
(Lys16-Asp23 and Lys28-Ala42) may adopt β-sheet secondary structures, and hence, have a higher beta-aggre-
gation propensity during fibrillation.
When a protein molecule binds to another biological polymer (protein) to form a complex, the subset of residues
in the interface that account for most of the protein binding free energy are called binding hot spots. Therefore,
Table 1. In silico-predicted ADME properties of DHA and ThT using FAF-Drugs 3 properties.
Properties DHA ThT
MW (molecular weight) 330.50 283.41
LogP (octanol-water partition coefficient) 5.84 4.66
LogSw (oral bioavailability scores based on solubility in water) −4.64 −4.67
tPSA (topological polar surface area, Å 2 ) 40.46 31.78
Number of rotatable bonds 14 2
Number of rigid bonds 6 16
Flexibility 0.70 0.11
Hydrogen Bond Donor (HBD) 2 0
Hydrogen Bond Acceptor (HBA) 2 2
Number of heavy atoms 24 20
Number of carbon atoms 22 17
Number of heteroatoms 2 3
* Number of lipinski violations
1 0
Solubility 3178.13 2665.28
FAF-Drugs 3 evaluated drug-like properties of ThT and DHA. FAF-Drugs 3 is a program for filtering large compound libraries prior to in silico
modeling studies. The tool can perform computational prediction of some ADME-tox properties (Adsorption, Distribution, Metabolism, Excretion,
and Toxicity) and offers free online services for calculation of important molecular properties (LogP, polar surface area, number of hydrogen bond
donors, and number of hydrogen bond acceptors), as well as prediction of the bioactivity score for the most important drug targets.
M. Hashimoto et al.
78
Table 2. Analyses of aggregation-prone amino acid residues in the Aβ 1-42 peptide sequence.
Amino acid
sequence
Leu Val Phe Ala Ala Glu Asp Val Gly Ser Asn Lys Gly Ala Ile Ile Gly Leu Met Val Gly Gly Val Val Ile Ala
1 -------------- 6 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42
FoldAmyloid 1
AGGRESCAN 2
ProA 3
1 FoldAmyloid [28], 2 AGGRESCAN [29], and 3 ProA [30] servers employ algorithms to predict amyloidogenic regions from protein amino acid se-
quence. The analyses are based on the amino acid physicochemical properties critical to protein aggregation. Color blocks indicate “common” amino
acid residues those are prone to aggregation, as analyzed by 1,2,3 Different servers.
Figure 3. Beta-aggregation propensity analysis of Aβ 1-42 . The red line indicates the region of the amino acid sequence
(which is shown as single-alphabet strip) that induces beta aggregation.
we analyzed the protein-protein interface interaction sites for the A1B1 dimer. Because amyloid fibrillation
starts with the interactions between at least two β strands, identifying these interaction sites is essential for un-
derstanding the nature of these sites, and whether DHA affects them, and the subsequent fibrillation. The Phe19
and Phe20 of the A1 monomer interacted with the Phe19 and Phe20 of the B1 monomer. The “Hot-point” server
principally considers the solvent accessibility and total contact potential of the interface residues. The output file
tabulates the interface residues with the highlighted hot spots and their features. The “hot-spot” with the highest
potential was located in Leu34 on chain B1 of 2BEG. The KFC2 showed that the interaction sites were at Phe19
and Phe19 of A1 and B1. Other amino acids from either monomeric chain involved in the inter-surface interac-
tions sites are highlighted in Table 3.
Pocketness is an important parameter that helps to determine the binding sites for ligands to their receptors.
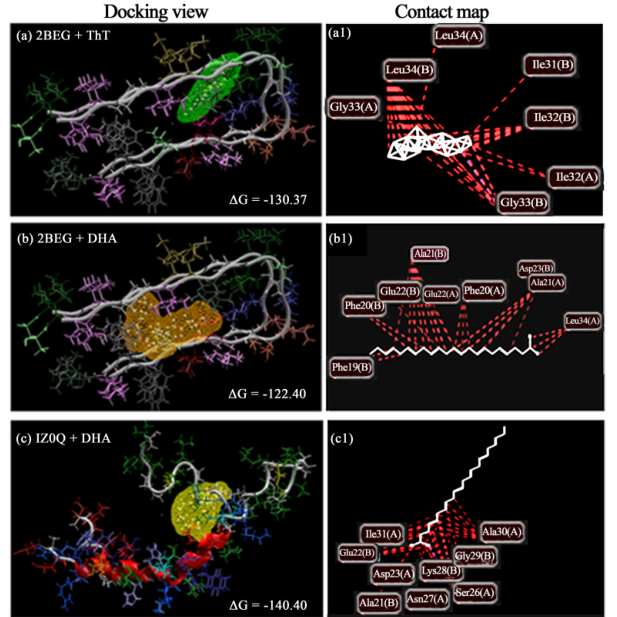
The 2BEG dimer or trimer showed ~5 or 6 pockets, whereas the 1Z0Q dimer displayed 4 pockets (Figure 4).
Docking performed by MVD provided the five best poses, and we obtained the corresponding MolDock score
values and other thermodynamically calculated values. The 3D structures of the best scoring dockings are shown
in Figures 5(a)-(c). Steric interactions of ThT and DHA with Aβ 1-42 (the receptor) were also evaluated with
MVD. Maps of the interactions of ThT with amino acid residues of the 2BEG dimer (A1B1) are shown in Fig-
ure 5(a1). The docking of DHA to dimeric Aβ 1-42 (A1B1 of 2BEG and A1A2 of 1Z0Q) and their interaction
maps are shown in Figure 5(b), Figure 5(b1), and Figure 5(c), Figure 5(c1), respectively. Using MVD, DHA
had a MolDock score of −122.40 Kcal/mol and −140.40 Kcal/mol when docked onto the 2BEG and 1Z0Q di-
mers, respectively. The ThT reference molecule had a MolDock score of −130.37 Kcal/mol when docked onto
the 2BEG dimer. Therefore, the binding of DHA to the 2BEG dimer was comparable with that of ThT, as
demonstrated by similar binding energies (Figure 5(b)).
Amino acids involved in the docking of DHA to the Aβ 1-42 dimer (2BEG, A1B1) and Aβ 1-42 trimer (1Z0Q,
A1A2) were visualized by contact maps of the interactions between atoms of DHA and the amyloid amino acid
M. Hashimoto et al.
79
Table 3. Analyses of momomer (A1)-monomer (B1) inter-surface interaction sites of Aβ 1-42 dimer (A1-B1).
Amino acid
sequence
Leu Val Phe Ala Ala Glu Asp Val Gly Ser Asn Lys Gly Ala Ile Ile Gly Leu Met Val Gly Gly Val Val Ile Ala
1 ------------------- 6 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42
PPISP 1
A1
B1
Hotpoint 2
A1
B1
KFC 3
A1
B1
1 Cons-PPISP is a consensus neural network method for predicting protein-protein interaction sites. Given the structure of a protein, cons-PPISP
will predict the residues that will likely form the binding site for another protein. The inputs to the neural network include position-specific se-
quence profiles and solvent accessibilities of each residue and its spatial neighbors. The neural network is trained on known structures of protein-
protein complexes [38]. 2 Hotpoint: Hotpoint, which predicts hot spots in protein interfaces using an empirical model. The empirical model in-
corporates a few rules consisting of occlusion from solvent and total knowledge-based pair potentials of residues [39]. 3 KFC2 (Knowledge-based
FADE and Contacts) server predicts binding “hot spots” within protein-protein interfaces by recognizing structural features indicative of impor-
tant binding contacts. The server analyzes several chemical and physical features surrounding an interface residue and predicts the classification
of the residue using a model trained on prior experimental data. Color blocks indicate “common” amino acid residues those acting at the inter-
surface interaction sites, as the Aβ 1-42 was analyzed by different servers.
Figure 4. Binding sites (pockets) analysis of Aβ 1-42 . One of the simplest ways to predict ligand binding sites is to identify
pocket-shaped regions on the protein surface. A binding sites or a pocket is defined as a space into which a small probe
can enter, but a large probe cannot. A pocket intrinsically has two arbitrary properties, size and depth. The GHECOM
analyzes these pockets using probe spheres. The radii of the probe spheres are assumed to correspond to the size and
depth of the pockets. These values can be adjusted to individual putative ligand molecule. The binding sites or pockets of
the dimer (A1B1) and trimer (A1B1C1) were determined by GHECOM, which detects grid-based pockets/binding sites
on the surface/interior of the protein [36]. The presence of binding sites was visualized using the 3D molecular viewer
Jmol [37]. The program also produces a graph of residue-based “pocketness” (right panel). (a1), (b1), (c1) number and
position of amino acids (represented as lines on the primary sequences) involved in each of the pocket of (a), (b) and (c)
of the left panel. For details please see Reference [36]. The individual color indicates the presence of distinct pockets.
M. Hashimoto et al.
80
Figure 5. The best-scored docking poses of ThT and DHA with the receptor (Aβ 1-42 ) by the MVD. (a) ThT docked onto
the 2BEG A1B1 dimer. (b) DHA docked onto the 2BEG A1B1dimer. (c) DHA docked on the 1Z0Q A1A2 dimer. Inte-
raction (binding) energies (∆G) are also shown. Contact (binding) maps of the interactions between atoms of ThT/DHA
and amino acid residues of the 2BEG/1Z0Q dimer are shown in the right panel (a1) (b1) and (c1). Contact maps were vi-
sualized by the “Ligand map” module of the MVD.
residues using MVD (Figure 5(b1) & Figure 5(c1), and Table 4). The amino acid residues of the A1B1 dimer
found to sterically interact with ThT were Ile32, Gly33, and Leu34 of A1 as well as Ile31, Ile32, Gly33, and
Leu34 of B1.
We also performed molecular docking of ThT and DHA using PatchDock to confirm the ability of DHA to
bind the 2BEG A1B1 dimer and the 2BEG A1B1C1 trimer (Figure 6). The algorithm used in PatchDock per-
forms rigid docking, with surface variability/flexibility implicitly addressed through liberal intermolecular pene-
tration. Geometric shape complementarity scores were similar for DHA and ThT when docked with the dimer
(Score: ThT = 3742; DHA = 3732). However, there was higher shape complementarity when DHA was docked
M. Hashimoto et al.
81
onto the trimer (Score: ThT = 3632; DHA = 4326). Approximate complex interface areas (receptor-ligand) were
also higher for DHA. ACEs for ThT and DHA were almost similar during docking onto the dimer; however, it
was slightly higher for ThT when docked onto the trimer (Table 5). These data suggest that DHA and ThT have
similar binding affinities.
Table 4. Patch docking of ThT and DHA with the 2BEG amyloid dimer (A1B1) and trimer (A1B1C1).
Docking (receptor vs. ligands) Score Area ACE
A1B1 dimer vs. ThT 3742 445.40 −248.98
A1B1 dimer vs. DHA 3732 496.70 −241.07
A1B1C1 trimer vs. ThT 3632 4.33.6 −219.71
A1B1C1 trimer vs. DHA 4326 542.10 −198.30
Abbreviations: Score, geometric shape complementarity score; Area, approximate interface area of the complex (receptor-ligand); ACE, atomic con-
tact energy; A1B1 dimer, generated by feeding the A1 and B1 model 1 of 2BEG to the Rosetta Server; A1B1C1 trimer, generated by feeding the A1,
B1, and C1 coordinate model 1 of 2BEG to the Rosetta Server; ThT, thioflavin T; DHA, docosahexaenoic acid.
Table 5. Summary of dockink of DHA and ThT on to NMR structures of Aβ 1-42 dimer structures (A1B1 of 2BEG and A1A2
of 1Z0Q).
Amino acid sequence Leu Val Phe Ala Ala Glu Asp Val Gly Ser Asn Lys Gly Ala Ile Ile Gly Leu Met Val Gly Gly Val Val Ile Ala
1 ------------------------ 6 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42
DHA
2BEG
A1
B1
1Z0Q
A1
B1
ThT 2BEG
A1
B1
When DHA and ThT were docked onto 2BEG dimer, they shared a common binding site at Ile31of A1 peptide. Both the 2BEG and 1Z0Q pdb had
common binding sites at Ala21 of A1 and Glu21 of B1 peptide. Color blocks at a given position (of the sequence) indicate common binding sites for
the ligand.
Figure 6. The best scoring results of the Patch Dock for ThT (a)-(c) and DHA (b)-(d) with the A1B1 dimer (a) (b) and the
A1B1C1 trimer (c) (d). Ligands (ThT and DHA) are represented by green sticks, and the receptors (dimer and trimer) are
shown as β-sheet ribbons with the amino acid side chains presented as sticks.
M. Hashimoto et al.
82
The ligand-docked receptor obtained from PatchDock was visualized using PyMOL as shown in Figure 6.
ThT bound with the interior regions of both the dimer and trimer (Figure 6(a) & Figure 6(c)), whereas DHA
was found to wrap around the β-sheet of the dimer (Figure 6(b) & Figure 6(d)).
4. Discussion
Alzheimer’s disease (AD) is one of the most common chronic dementia-related diseases worldwide, and the
number of AD patients is increasing every year. Currently, only two medications (cholinesterase inhibitors and
an NMDA receptor antagonist) are approved by the United States Food and Drug Administration [44] to treat
the cognitive symptoms (memory loss and confusion) of AD. However, these medications cannot cure AD or
arrest its progression. Vitamins or other supplements, such as omega-3 fatty acids [45], and various herbal mix-
tures are also widely promoted as preparations that may support cognitive health or prevent or delay AD. The
NIH panel concluded that there is somewhat stronger data—but not definitive evidence—that omega-3 fatty ac-
ids in fish oil may help prevent cognitive decline. The basis of such conclusion might relate to the fact that the
concentration of omega-3 polyunsaturated docosahexaenoic acid (DHA, C22:6, n-3) is significantly reduced in
the brains of AD patients [21] [22]. We also reported on the beneficial effects of dietary omega-3 polyunsatu-
rated fatty acid supplementation on age-related cognitive decline in elderly Japanese patients with very mild
dementia [46]. Therefore, DHA supplementation may decelerate the cognitive symptoms of AD.
We previously showed that chronic administration of DHA significantly decreased the amyloid burden in the
brains of AD model rats [14]. DHA also inhibited the in vitro amyloid fibrillations of Aβ 1-40 [16], Aβ 1-42 [10],
and Aβ 25-35 [18], indicating that neurotoxicity convened by these amyloid species would be inhibited by the
DHA. Moreover, we found that Aβ 1-42 -induced toxicity of SH-S5Y5 cells was reduced by DHA [10], andour
previous transmission electron microscopy, laser-scanning fluorescence microscopy, and thioflavin T fluoros-
pectroscopy studies clearly indicated that DHA resists fibrillogenesis by some unknown mechanism [10] [16]
[18]. Despite strong evidences that soluble forms of Aβ are toxic at synapses, the exact forms of the toxic spe-
cies remain to be determined. There is evidence for toxicity of both low molecular weight dimers and trimers of
Aβ [47]-[50]. Using Western blot analysis and/or polyacrylamide gel electrophoresis, we have also shown that
DHA inhibits the fibrillation of dimeric/trimeric and oligomeric Aβs [10]; thus, it ultimately decreases the elon-
gation process of amyloid fibrillation leading to matured fibers. Finally, the anti-Aβ fibrillation effect of DHA
was further supported by the reductions of Aβ-induced apoptosis and neurodegeneration [10] in DHA-treated
cell culture. These in vivo/in vitro experimental data strongly suggest that DHA prevents amyloid peptides from
forming toxic species that ultimately deteriorate memory and other behavioral aspects in AD patients.
These findings also demonstrate that Aβ peptides are important drug targets, and designing drugs that disrupt
the formation of Aβ-Aβ inter-surface interaction sites is particularly important for preventing Aβ fiber formation.
Therefore, we performed the present in silico study to support the experimentally proven ameliorative effects of
DHA on the Aβ-fibrillation. Accordingly, we modeled the 3D structure of DHA and used docking simulation to
determine whether DHA binds to Aβ peptides and to predict the affinity between the ligand (DHA) and the re-
ceptor (Aβ). During docking simulations with both MVD and PatchDock, we used ThT as a positive control to
verify whether docking would yield ligand geometry similar to that of DHA when binding to Aβ peptides.
Using MVD, DHA had a MolDock score of −122.4 Kcal/mol with 2BEG, and −140.40 Kcal/mol with 1Z0Q,
whereas ThT had a MolDock score of −130.37 Kcal/mol. These results suggest considerable binding of DHA
with the dimeric Aβ 1-42 molecules. This might explain the reduced amyloid burden observed in the brains of
DHA-administered AD model rats [16] and concurrent inhibition of in vitro fibril formation [10]. Monomer-
monomer inter-surface interaction site analyses revealed that Phe19 (A1) and Phe19 (B1) were the common
amino acid residues, which acted as monomer-monomer inter-surface interaction sites (Table 3). Other amino
acid residues, including Phe20 (A1), Phe20 (B1), Ala21 (A1), Asp23 (A1), Gly25 (A1), Lys28 (B1), Ile32 (B1),
Gly33 (A1), Leu34 (B1), Met (A1), Val36 (A1), Val36 (B1), Gly37 (A1), Gly37 (B1), Gly38 (A1), Val39 (B1),
Val 40 (A1), and Ile41 (B1) also acted as inter-surface interaction sites (Table 3). These inter-surface interaction
sites might have been targeted by the DHA, as indicated by the interactions of DHA with these amino acid re-
gions, particularly, those of Phe19, Ala21, Asp23, and Leu34. Notably, DHA also interacted with Ala21, Glu22,
Asp23, and Ser26-Ile31 of 1Z0Q, which is in an α -helix random coil conformation rather than the β -sheet con-
formation that is the prerequisite for the propagation of fibrillation. As shown in Table 2, the amino acid resi-
dues 24 - 29 and 39 - 42 are the most aggregation-sensitive regions of Aβ 1-42 . All these results suggest that DHA
M. Hashimoto et al.
83
interacts with these aggregation-prone monomer-monomer inter-surface interaction sites. Moreover, ThT
docked with 2BEG in a similar manner to DHA and bound to Ile31, Ile32, Gly33, and Leu34.
In the PatchDock system, the docking of DHA and ThT were similar, as indicated by the almost similar
docking score. However, when they were docked by PatchDock onto the Aβ 1-42 trimer, DHA displayed a greater
binding affinity (score) than ThT. When the PatchDock receptor-ligand was visualized with PyMOL (Figure 6),
an interesting feature was obtained: DHA spirally wrapped the dimer, whereas ThT bound to the interior of the
dimer. Notably, DHA and ThT both bound to the predicted binding pockets (Figure 4). The binding interaction
between the ligand molecule (DHA) and receptor (Aβ 1-42 ) has a significant role in determining the anti-amyloid
activity. Therefore, the binding potential of DHA in this computational study supports the results of our previous
in vivo and in vitro biological studies showing that DHA decreases in vitro amyloid fibrillation and amyloid
brain burden in a rat AD model, with concurrent amelioration of Aβ-induced memory loss [10] [14] [16] [18].
AD drug discovery has been a very lengthy and costly process, and the prevention of AD remains a burning is-
sue worldwide. Polyunsaturated fatty acids, including DHA, have been continually investigated over the last
several years as potential drug candidates with minimal side effects in experimental animal models of AD and/or
AD human patients. Using a targeted and more specific approach for future high-throughput screens, such as
those based on the results of similar in silico experiments performed here, we may be able to more rapidly iden-
tify novel AD drugs.
5. Conclusion
Computational drug design is a promising method for discovering many more drugs in the coming years. Here
we demonstrated that molecular docking of DHA is consistent with its anti-amyloid effects, in particular, on in
vitro fibrillation and in vivo Aβ-induced memory impairments of AD model rats. Our study further reaffirms that
DHA is important for the structure and biological function of Aβs. We also showed that scoring the docking
values gives the best prediction for ligand interactions with proteins. Finally, our in silico results provide com-
pelling evidence for the utility of DHA as a preventive medication for neurodegenerative diseases such as
Aβ 1-42 -induced AD. Further studies on DHA-like molecules hold promise for the development of anti-amyloid
drugs.
Acknowledgements
This work was supported in part by a Grant-in-Aid for Scientific Research (C) from the Ministry of Education
Culture, Sports, Science and Technology, Japan (26500008 to M.H.). We also gratefully acknowledge the MVD
authority for providing us with the free academic version.
Conflict of Interest
The authors declare no-conflict of interest.
References
[1] Rajan, K.B., Wilson, R.S., Weuve, J., Barnes, L.L. and Evans, D.A. (2015) Cognitive Impairment 18 Years before
Clinical Diagnosis of Alzheimer Disease Dementia. Neurology.
[2] Sindi, S., Mangialasche, F. and Kivipelto, M. (2015) Advances in the Prevention of Alzheimer’s Disease. F1000Prime
Reports, 7, 50.
http://dx.doi.org/10.12703/P7-50[3] Bacanu, S.A., Devlin, B., Chowdari, K.V., DeKosky, S.T., Nimgaonkar, V.L. and Sweet, R.A. (2005) Heritability of
Psychosis in Alzheimer Disease. American Journal of Geriatric Psychiatry, 13, 624-627.
http://dx.doi.org/10.1097/00019442-200507000-00011[4] Selkoe, D.J. (1991) The Molecular Pathology of Alzheimer’s Disease. Neuron, 6, 487-498.
http://dx.doi.org/10.1016/0896-6273(91)90052-2[5] Citron, M. (2010) Alzheimer’s Disease: Strategies for Disease Modification. Nature Reviews Drug Discovery, 9, 387-
398.
http://dx.doi.org/10.1038/nrd2896[6] De Strooper, B., Vassar, R. and Golde, T. (2010) The Secretases: Enzymes with Therapeutic Potential in Alzheimer
Disease. Nature Reviews Neuroscience, 6, 99-107.
http://dx.doi.org/10.1038/nrneurol.2009.218[7] Golde, T.E., Schneider, L.S. and Koo, E.H. (2011) Anti-A β Therapeutics in Alzheimer’s Disease: The Need for Apa-
M. Hashimoto et al.
84
radigm Shift. Neuron, 69, 203-213.
http://dx.doi.org/10.1016/j.neuron.2011.01.002[8] Huang, Y. and Lennart Mucke, L. (2012) Alzheimer Mechanisms and Therapeutic Strategies. Cell, 148, 1204-1222.
http://dx.doi.org/10.1016/j.cell.2012.02.040[9] Palop, J.J. and Mucke, L. (2010) Amyloid-Beta-Induced Neuronal Dysfunction in Alzheimer’s Disease: From Syn-
apses toward Neural Networks. Nature Neuroscience, 13, 812-818.
http://dx.doi.org/10.1038/nn.2583[10] Hossain, S., Hashimoto, M., Katakura, M., Miwa, K., Shimada, T. and Shido, O. (2009) Mechanism of Docosahexae-
noicacid-Induced Inhibition of in Vitro Aβ 1-42 Fibrillation and Aβ 1-42 -Induced Toxicity in SH-S5Y5 Cells. Journal of
Neurochemistry, 111, 568-579.
http://dx.doi.org/10.1111/j.1471-4159.2009.06336.x[11] Goedert, M. and Spillantini, M.G. (2006) A Century of Alzheimer’s Disease. Science, 314, 777-781.
http://dx.doi.org/10.1126/science.1132814[12] Song, Y., Chen, X., Wang L.Y., Gao, W. and Zhu, M.J. (2013) Rho Kinase Inhibitor Fasudil Protects Against β-Amy-
loid-Induced Hippocampal Neurodegeneration in Rats. CNS Neuroscience & Therapeutics, 19, 603-610.
http://dx.doi.org/10.1111/cns.12116[13] Mamun, A.A., Hashimoto, M., Katakura, M., Matsuzaki, K., Hossain, S., Arai, H. and Shido, O. (2014) Neuroprotec-
tive Effect of Madecassoside Evaluated Using Amyloid Β1-42-Mediated in Vitro and in Vivo Alzheimer’s Disease
Models. International Journal of Indigenous Medicinal Plants, 47, 1669-1682.
[14] Hashimoto, M., Hossain, S., Shimada, T., Sugioka, K., Yamasaki, H., Fujii, Y., Ishibashi, Y., Oka, J. and Shido, O.
(2002) Docosahexaenoic Acid Provides Protection from Impairment of Learning Ability in Alzheimer’s Disease Model
Rats. Journal of Neurochemistry, 81, 1084-1091.
http://dx.doi.org/10.1046/j.1471-4159.2002.00905.x[15] Hashimoto, M., Hossain, S. and Shido, O. (2005) Docosahexaenoic Acid-Induced Amelioration on Impairment of
Memory Learning in Amyloid Beta-Infused Rats Relates to the Decreases of Amyloid Beta and Cholesterol Levels in
Detergent-Insoluble Membrane Fractions. Biochimica et Biophysica Acta, 1738, 91-98.
http://dx.doi.org/10.1016/j.bbalip.2005.11.011[16] Hashimoto, M., Shahdat, H.M., Yamashita, S., Katakura, M., Tanabe, Y., Fujiwara ,H., Gamoh, S., Miyazawa, T., Arai
H., Shimada, T. and Shido, O. (2008) Docosahexaenoic Acid Disrupts in Vitro Amyloid Beta(1-40)Fibrillation and
Concomitantly Inhibits Amyloid Levels in Cerebral Cortex of Alzheimer’s Disease Modelrats. Journal of Neurochemi-
stry, 107, 1634-1646.
http://dx.doi.org/10.1111/j.1471-4159.2008.05731.x[17] Kubo, T., Nishimura, S., Kumagae, Y. and Kaneko, I. (2002) In Vivo Conversion of Racemized β -Amyloid ([D-Ser 26 ]
A β 1-40 ) to Truncated and Toxic Fragments ([D-Ser 26 ]A β 25-35/40 ) and Fragment Presencein the Brains of Alzheimer’s Pa-
tients. Journal of Neuroscience Research, 70, 474-483.
http://dx.doi.org/10.1002/jnr.10391[18] Hashimoto, M., Shahdat, H.M., Katakura, M., Tanabe, Y., Gamoh, S., Miwa, K., Shimada, T. and Shido, O. (2009)
Effects of Docosahexaenoic Acid on in Vitro Amyloid Beta Peptide 25-35 Fibrillation. Biochimica et Biophysica Acta,
1791, 289-296.
http://dx.doi.org/10.1016/j.bbalip.2009.01.012[19] Soderberg, M., Edlund, C., Kristensson, K. and Dallner, G. (1991) Fatty Acid Composition of Brain Phospholipidsin
Aging and in Alzheimer’s Disease. Lipids, 26, 421-425.
http://dx.doi.org/10.1007/BF02536067[20] Prasad, M.R., Lovell, M.A., Yatin, M., Dhillon, H. and Markesbery, W.R. (1998) Regional Membrane Phospholipidal-
terations in Alzheimer’s Disease. Neurochemical Research, 23, 81-89.
http://dx.doi.org/10.1023/A:1022457605436[21] Morris, M.C., Evans, D.A., Bienias, J.L., Tangney, C.C., Bennett, D.A., Wilson, R.S., Aggarwal, N. and Schneider, J.
(2003) Consumption of Fish and n-3 Fatty Acids and Risk of Incident Alzheimer Disease. Archives of Neurology, l60,
940-946.
http://dx.doi.org/10.1001/archneur.60.7.940[22] Huang, T.L., Zandi, P.P., Tucker, K.L., Fitzpatrick, A.L., Kuller, L.H., Fried, L.P., Burke, G.L. and Carlson, M.C.
(2005) Benefits of Fatty Fish on Dementia Risk Are Stronger for Those without APOE Epsilon4. Neurology, 65, 1409-
1414.
http://dx.doi.org/10.1212/01.wnl.0000183148.34197.2e[23] Mamun, A.A., Hashimoto, M., Hossain, S., Katakura, M., Matsuzaki, K., Arai, H. and Shido, O. (2015) Confirmation
of the Experimentally-Proven Therapeutic Utility of Madecassoside in an Aβ 1-42 InfusionRat Model of Alzheimer’s
Disease by in Silico Analyses. Advances in Alzheimer’s Disease, 4, 37-44.
http://dx.doi.org/10.4236/aad.2015.42005[24] Schüttelkopf, A.W. and van Aalten, D.M.F. (2004) PRODRG—A Tool for High-Throughput Crystallography of Pro-
tein-Ligand Complexes. Acta Crystallographica, D60, 1355-1363.
http://dx.doi.org/10.1107/S0907444904011679[25] Lipinski, C.A., Lombardo, F., Dominy, B.W. and Feeney, P.J. (2001) Experimental and Computational Approaches to
Estimate Solubility and Permeability in Drug Discovery and Development Settings. Advanced Drug Delivery Reviews,
46, 3-26.
http://dx.doi.org/10.1016/S0169-409X(00)00129-0[26] Ertl, P., Rohde, B. and Selzer, P. (2000) Fast Calculation of Molecular Polar Surface Area as a Sum of Fragment-Based
Contributions and Its Application to the Prediction of Drug Transport Properties. Journal of Medicinal Chemistry, 43,
3714-3717.
http://dx.doi.org/10.1021/jm000942eM. Hashimoto et al.
85
[27] Daylight Chemical Information Systems Inc. (2007) SMARTS—A Language for Describing Molecular Patterns. Aliso
Viejo.
[28] Garbuzynskiy, S.O., Lobanov, M.Y. and Galzitskaya, O.V. (2010) Fold Amyloid: A Method of Prediction Ofamyloi-
dogenic Regions from Protein Sequence. Bioinformatics (Oxford, England), 26, 326-332.
http://dx.doi.org/10.1093/bioinformatics/btp691[29] Conchillo-Sole, O., de Groot, N.S., Aviles, F.X., Vendrell, J., Daura , X. and Ventura, S. (2007) AGGRESCAN:
Aserver for the Prediction and Evaluation of “Hot Spots” of Aggregation in Polypeptides. BMC Bioinformatics, 8, 65.
http://dx.doi.org/10.1186/1471-2105-8-65[30] Fang, Y., Gao, S., Ta, D., Middaugh, C.R. and Fang, J. (2013) Identification of Properties Important to Protein Aggre-
gation Using Feature Selection. BMC Bioinformatics, 14, 314.
http://dx.doi.org/10.1186/1471-2105-14-314[31] Rousseau, F., Schymkowitz, J. and Serrano, L. (2006) Protein Aggregation and Amyloidosis: Confusion of the Kinds?
Current Opinion in Structural Biology, 16, 118-126.
http://dx.doi.org/10.1016/j.sbi.2006.01.011[32] Fernandez-Escamilla A.M., Rousseau, F., Schymkowitz, J. and Serrano, L. (2004) Prediction of Sequence-Dependent
and Mutational Effects on the Aggregation of Peptides and Proteins. Nature Biotechnology, 22, 1302-1306.
http://dx.doi.org/10.1038/nbt1012[33] Linding, R., Schymkowitz, J., Rousseau, F., Diella, F. and Serrano, L. (2004) A Comparative Study of the Relationship
between Protein Structure and Beta-Aggregation in Globular and Intrinsically Disordered Proteins. Journal of Molecu-
lar Biology, 342, 345-353.
http://dx.doi.org/10.1016/j.jmb.2004.06.088[34] Lyskov, S. and Gray, J.J. (2008) The RosettaDock Server for Local Protein-Protein Docking. Nucleic Acids Research,
36, W233-W238.
http://dx.doi.org/10.1093/nar/gkn216[35] Hossain, S., Hashimoto, M., Katakura, M., Mamun, A.A. and Shido, O. (2015) Medicinal Value of Asiaticoside for
Alzheimer’s Disease as Assessed Using Single-Molecule-Detection Fluorescence Correlation Spectroscopy, Laser-
Scanning Microscopy, Transmission Electron Microscopy, and in Silico Docking. BMC Complementary and Alterna-
tive Medicine, 15, 118.
http://dx.doi.org/10.1186/s12906-015-0620-9[36] Kawabata, T. (2010) Detection of Multi-Scale Pockets on Protein Surfaces Using Mathematical Morphology. Proteins,
78, 1195-1221.
http://dx.doi.org/10.1002/prot.22639[37] Jmol: An Open-Source Java Viewer for Chemical Structures in 3D.
http://www.jmol.org/[38] Chenm, H.-L. and Zhou, H.-X. (2005) Prediction of Interface Residues in Protein-Protein Complexes by a Consensus
Neural Network Method: Test against NMR Data. Proteins, 61, 21-35.
http://dx.doi.org/10.1002/prot.20514[39] Tuncbag, N., Gursoy, A. and Keskin, O. (2009) Identification of Computational Hotspots in Protein Interfaces: Com-
bining Solvent Accessibility and Inter-Residue Potentials Improves the Accuracy. Bioinformatics, 25, 1513-1520.
http://dx.doi.org/10.1093/bioinformatics/btp240[40] Zhu, X. and Mitchell, J.C. (2011) KFC2: A Knowledge-Based Hot Spot Prediction Method Based on Interface Solva-
tion, Atomic Density and Plasticity Features. Proteins, 79, 2671-2683.
http://dx.doi.org/10.1002/prot.23094[41] Thomsen, R. and Christensen M.H. (2006) MolDock: A New Technique for High-Accuracy Molecular Docking.
Journal of Medicinal Chemistry, 49, 3315-3321.
http://dx.doi.org/10.1021/jm051197e[42] Duhovny, D., Nussinov, R., Wolfson, H.J., et al. (2002) Efficient Unbound Docking of Rigid Molecules. Proceedings
of the 2nd Workshop on Algorithms in Bioinformatics (WABI), Vol. 2452, Rome, 17-21 September 2002, 185-200.
http://dx.doi.org/10.1007/3-540-45784-4_14[43] The PyMOL (1.7.4.4 Edu) Molecular Graphics System, Version 1.8 Schrödinger, LLC.
[44]
http://www.alz.org/research/science/alzheimers_disease_treatments.asp#approved[45]
http://www.alz.org/professionals_and_researchers_alternative_treatments_.asp#omega[46] Hashimoto, M., Yamashita, K. and Kato, S. (2012) Beneficial Effects of Dietary Omega-3 Polyunsaturated Fatty Acid
Supplementation on Age-Related Cognitive Decline in Elder Japanese with Very Mild Dementia: A 2-Year Rando-
mized, Double-Blind, Placebo-Controlled Trial. The Journal of Aging Research & Clinical Practice, 1, 193-201.
[47] Jin, M., Shepardson, N., Yang, T., Chen, G., Walsh, D. and Selkoe, D. (2011) Soluble Amyloid Beta-Protein Dimers
Isolated from Alzheimer Cortex Directly Induce Tau Hyperphosphorylation and Neuritic Degeneration. Proceedings of
the National Academy of Sciences of the United States of America, 108, 5819-5824.
http://dx.doi.org/10.1073/pnas.1017033108[48] Masters, C. and Selkoe, D. (2012) Biochemistry of Amyloid β-Protein and Amyloid Deposits in Alzheimer Disease.
Cold Spring Harbor Perspectives in Medicine, 2, a006262.
http://dx.doi.org/10.1101/cshperspect.a006262[49] Mc Donald, J., Savva, G., Brayne, C., Welzel, A., Forster, G., Shankar, G., Selkoe, D., Ince, P. and Walsh, D. (2010)
Medical Research Council Cognitive F and Ageing S. The Presence of Sodium Dodecyl Sulphatestable Abeta Dimers
M. Hashimoto et al.
86
Is Strongly Associated with Alzheimer-Type Dementia. Brain, 133, 1328-1341.
http://dx.doi.org/10.1093/brain/awq065[50] Shankar, G.M., Li, S., Mehta, T.H., Garcia-Munoz, A., Shepardson, N.E., Smith, I., Brett, F.M., Farrell, M.A., Rowan,
M.J., Lemere, C.A., et al. (2008) Amyloid-Beta Protein Dimers Isolated Directly from Alzheimer’s Brains Impair
Synaptic Plasticity and Memory. Nature Medicine, 14, 837-842.
http://dx.doi.org/10.1038/nm1782Abbreviations Used
AD: Alzheimer’s Disease;
A β : Amyloid Beta Peptide;
RCSB: Research Collaboratory for Structural Bioinformatics;
PBD: Protein Data Bank;
MVD: Molegro Virtual Docker;
ThT: Thioflavin T;
DHA: Docosahexaenoic Acid

 Computational Analyses of Docosahexaenoic
Computational Analyses of Docosahexaenoic